Research Focus |
|
类器官 2024-03-19 19:07:45 浏览次数:556 | |
| 类器官 Organoids nature 类器官是简单的基于组织工程细胞的体外模型,概括了相应体内组织的复杂结构和功能的许多方面。它们可以被解剖和询问,用于人体组织发育、再生和修复的基本机制研究,也可以用于诊断、疾病建模、药物发现和个性化医学。类器官来源于多能干细胞或组织驻留干细胞(胚胎或成体),或来自健康或患病组织(如肿瘤)的祖细胞或分化细胞。迄今为止,已经报道了许多支持类器官培养和生长、增殖、分化和成熟的类器官工程策略。 本初级读本强调了选择和开发这些材料和方法以控制细胞/组织生态位的基本原理;以及因此工程类有机物的结构和功能。我们还讨论了产生强大类器官的关键考虑因素,如与细胞分离和播种、基质和可溶性因子选择、物理线索和整合有关的因素。还概述了类器官群落中数据质量、再现性和沉积的一般标准。最后,我们最后阐述了类有机物在不同应用中的局限性,以及未来几年类有机物工程的关键优先事项。 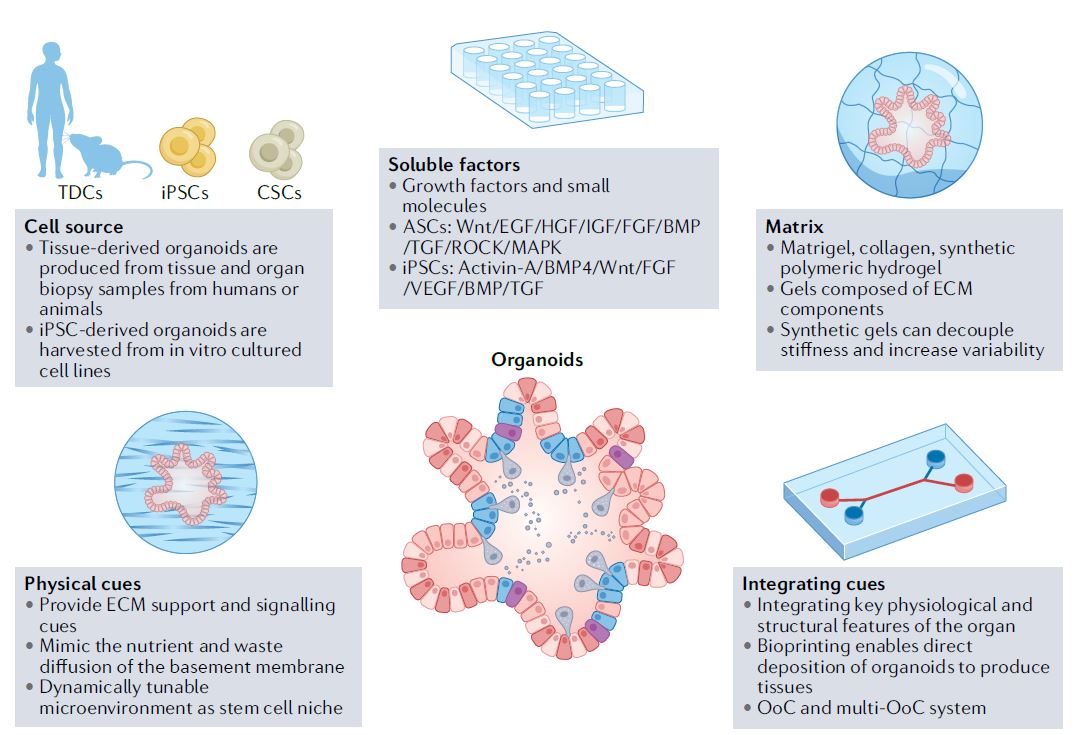 干细胞在通过细胞更新、迁移、分化和凋亡维持器官大小、结构和功能方面至关重要。干细胞位于一个定义的微环境中,通常被称为干细胞生态位,以调节干细胞的命运。鉴于这些环境线索的重要性,已经有许多组织工程尝试在体外模拟干细胞生态位,以实现对细胞-细胞和细胞-基质相互作用的高度时空控制,并使用工程水凝胶和微型设备复制机械化学线索。1977年,Matrigel从小鼠肉瘤肿瘤中提取出来,用于支持体外细胞培养。Matrigel是一种基底膜细胞外基质(ECM),含有ECM成分和生长因子的独特混合物。Matrigel后来被证明可以使乳腺上皮细胞三维生长,并与乳蛋白分泌形成管腔6,在存在组织特异性生长因子混合物的情况下,嵌入Matrigel中的成年肠道干细胞也被证明可以自组织成3D隐窝-绒毛结构。一个多世纪以来,类器官研究与3D细胞培养、干细胞和组织工程交织在一起,在定义、标准和范围上存在各种争论。 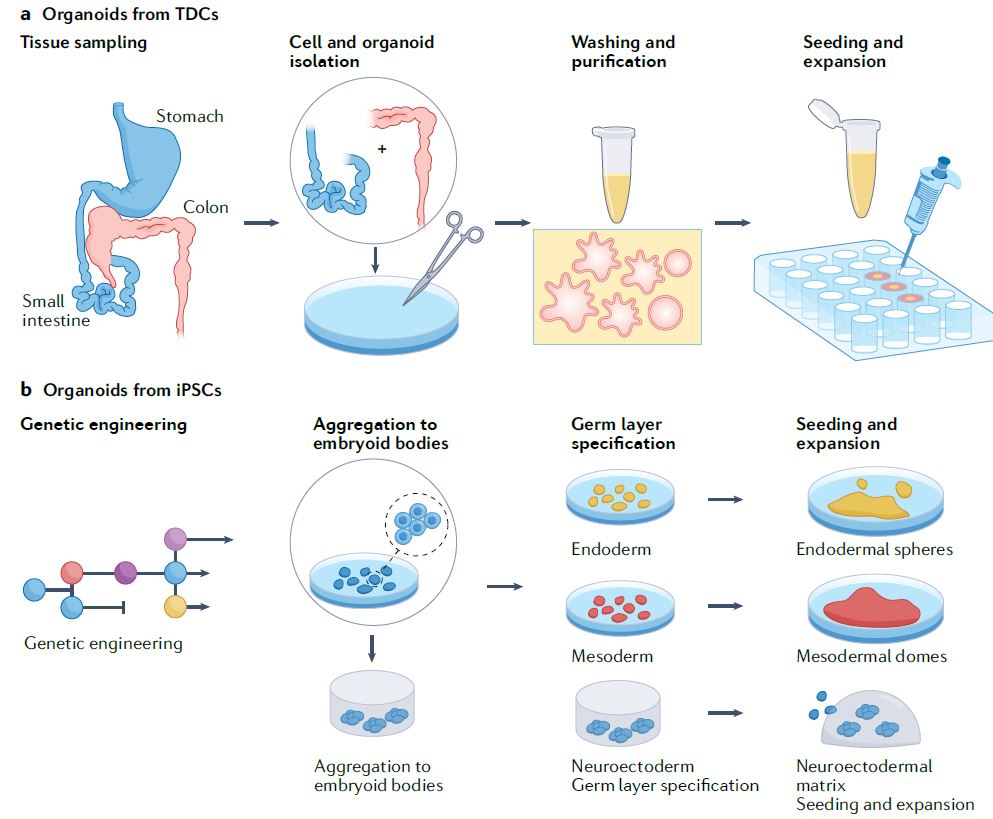 类器官是一种自组织的3D组织,通常来源于干细胞(多能干细胞、胎儿或成人),模仿器官的关键功能、结构和生物复杂性。包含类器官的细胞可衍生自诱导多能干细胞(iPSC)或组织衍生细胞(TDCs),包括正常干/祖细胞、分化细胞和癌症细胞。与传统的2D培养物和动物模型相比,类器官培养物能够在模型中实现患者特异性,同时在体外概括体内组织样结构和功能。类器官培养物比动物模型更易于操作和深入的生物学研究。因此,类器官培养物已被用于广泛的应用,包括药物发现、个性化伴随诊断15和细胞治疗。 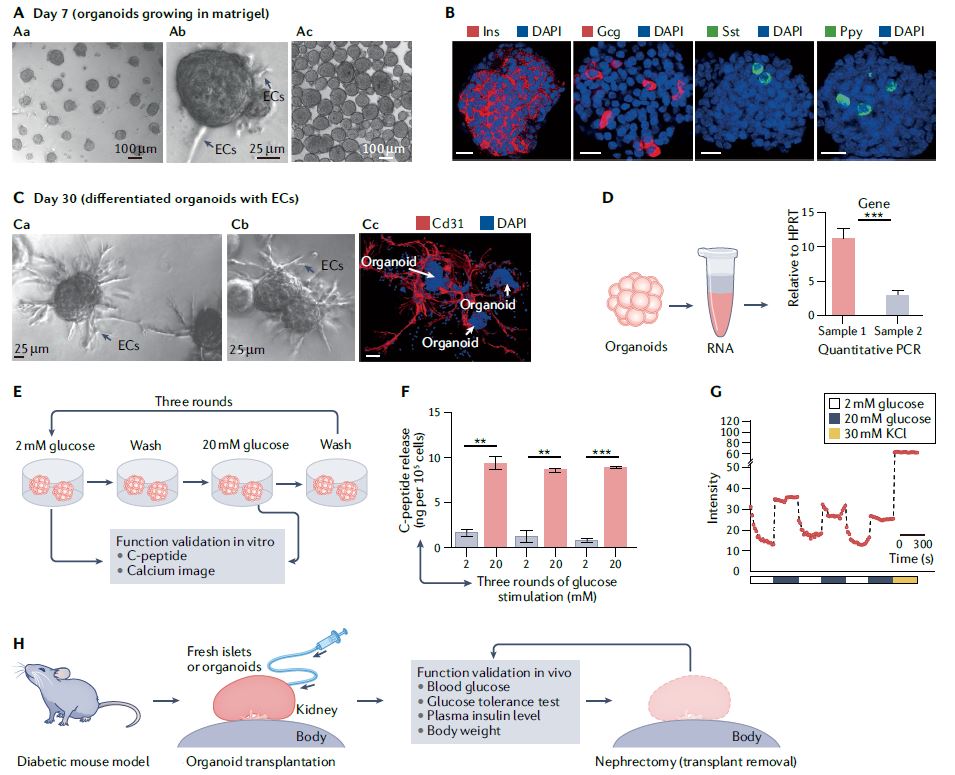 展望未来,趋势是开发更复杂的模型,尽可能忠实地概括体内结构和功能,包括随着时间的推移概括的细胞类型、组织结构、可测量的分子事件和表型功能。与其只关注最突出的标志物或功能测定,还应进行天然组织的结构基准测试。在肝细胞类器官的例子中,肝细胞功能得以保留,但肝组织结构与肝细胞排列成绳索的天然组织不匹配。类似地,类器官,如胰腺癌或结肠癌癌症类器官各向同性生长,形成囊肿,而不是它们在天然组织中形成的管状结构。为了获得更复杂的功能,具有多细胞和多组织结构的类器官将是重要的,特别是在研究细胞-细胞相互作用的背景下。沿着这条脉络,芯片上的组装体和器官也变得越来越复杂,并被更广泛地采用。 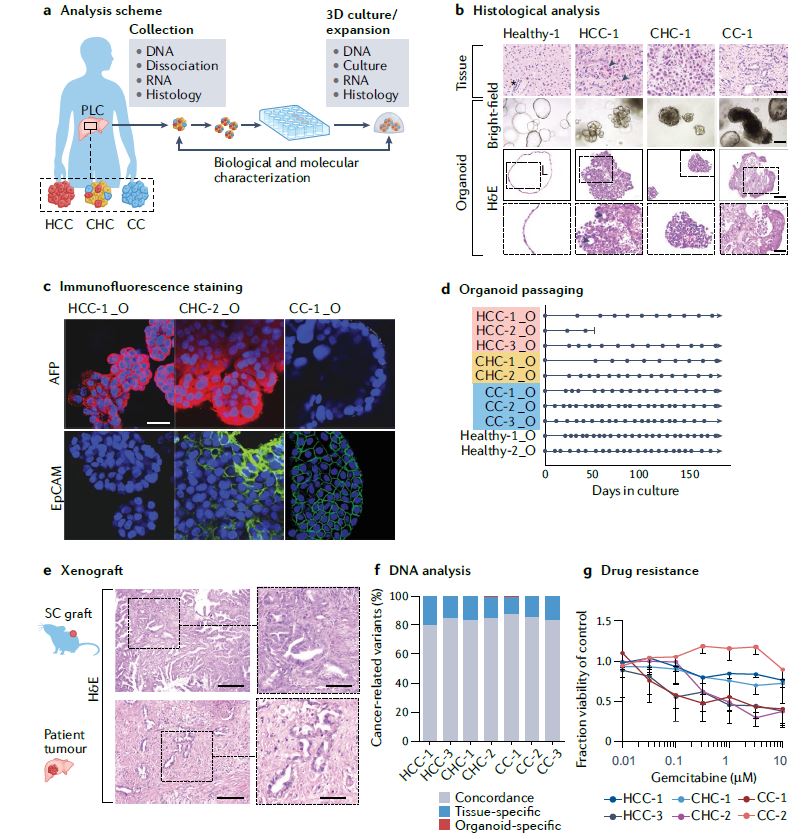 另一方面,工程师的方法是追求由驱动感兴趣的复杂细胞或组织功能的最小功能模块定义的更简单的还原论模型,研究发育或修复中的机械生物学原因,或开发用于高通量筛选的强大系统。基本前提是,复杂的生物功能是通过有限数量的功能模块的协调操作来执行的,每个功能模块由一小组分子描述,以及驱动中尺度(亚细胞或细胞间组织/多细胞)物理属性变化的化学反应与特定时空阶段/阶段/步骤中感兴趣的功能相关联的结构。例如,肝脏中的胆管表现出每小时一次的扩张和收缩周期。为了高分辨率地研究引起收缩的事件,仅在相邻肝细胞的整个调节机制的背景下直接研究形成胆管的相邻肝细胞区域。人们可以选择创建一个比由最小功能模块操作的结构大得多的涉及胆管细胞的结构,但该模型将是嘈杂和昂贵的。每个功能模块相互耦合,可以在不同的长度尺度上一起或独立建模。简单的还原论模型有助于对组织形态发生事件(如缺陷)的高分辨率机制理解。通过使用Matrigel进行微图案化和支持3D细胞生长,在几何上限制初始2D种子图案的大小和3D形成,能够诱导组织样神经管的形态发生和生产高度可再生的神经管。这也允许识别神经管折叠的机制并对神经管缺陷进行建模。在另一个例子中,均匀细胞球体中的对称性破坏和Paneth细胞的出现是肠道类器官形成早期的关键事件。其机制最近被阐明:对称性破坏是由机械转导器YAP1的瞬时激活引起的,其诱导NOTCH-DLL1横向抑制事件。此后,YAP1的激活通过在水凝胶支架中应用几何约束并产生均匀且可重复的肠道微组织来控制3。类器官可以通过在2.5D培养物中减少第三维度来限制。2.5D培养降低了典型类器官的深度驱动的可变性:缺氧核心的扩散限制、药物/转染剂的可及性有限以及成像透明度受阻。第三维限制的典型例子包括在弯曲或图案化的表面上培养细胞,平坦或受约束的细胞构建体,并在高融合度下将ECM覆盖在平坦的细胞单层上,这将把细胞向上拉,迫使更多的细胞-细胞相互作用采用3D细胞形态。胶原夹层中的肝细胞具有足够的接触面积,以获得极性并形成胆管管腔,该胆管管腔在没有3D网络的情况下以与体内相同的周期收缩,与天然组织相比更宽且胆汁淤积。这种基于细胞的模型能够高分辨率地将胆管收缩的机制分解为步骤,并理解调节相变的分子机制。我们还将设想更多基于CRISPR编辑的细胞的工程类器官模型用于疾病建模,尽管这些细胞和模型是合成的。 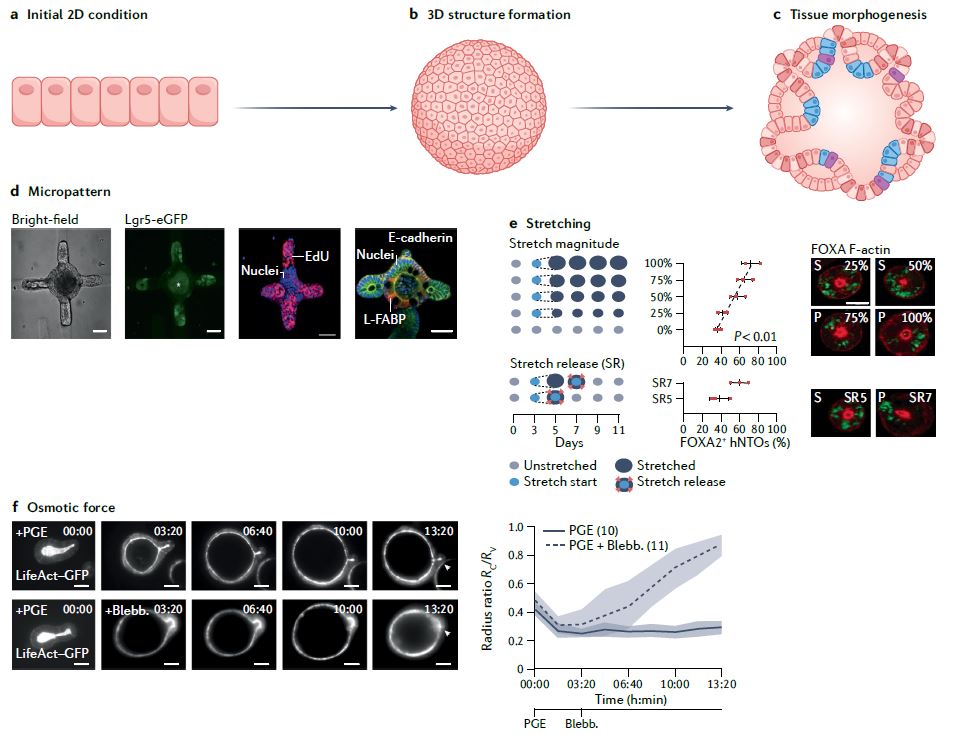 除了在创建更具生理相关性、更稳健、更易于使用的类器官模型方面的技术进步外,我们还设想在应用中看到更大的影响。在过去的二十年里,尽管有人讨论过取代动物试验,但这些努力尚未导致具体行动。然而,这种情况正在迅速发生变化,监管方面现在在多个方面建立了严格的阻止措施。能够概括体内复杂生理功能的类器官也增强了人们的信心,即新的替代方法现在是可行的选择。将对人类类器官的动物研究结果进行更多的推断,以更好地了解人类生物学和病理生理学。我们设想广泛采用类器官作为细胞源,用于细胞治疗、再生医学、体外诊断和药物发现。 Organoids are simple tissue-engineered cell-based in vitro models that recapitulate many aspects of the complex structure and function of the corresponding in vivo tissue. They can be dissected and interrogated for fundamental mechanistic studies on development, regeneration and repair in human tissues, and can also be used in diagnostics, disease modelling, drug discovery and personalized medicine. Organoids are derived from either pluripotent or tissue-resident stem (embryonic or adult) or progenitor or differentiated cells from healthy or diseased tissues, such as tumours. To date, numerous organoid engineering strategies that support organoid culture and growth, proliferation, differentiation and maturation have been reported. This Primer highlights the rationale underlying the selection and development of these materials and methods to control the cellular/tissue niche; and therefore, the structure and function of the engineered organoid. We also discuss key considerations for generating robust organoids, such as those related to cell isolation and seeding, matrix and soluble factor selection, physical cues and integration. The general standards for data quality, reproducibility and deposition within the organoid community are also outlined. Lastly, we conclude by elaborating on the limitations of organoids in different applications, and the key priorities in organoid engineering for the coming years. Stem cells are critical in maintaining organ size, structure and function through cellular renewal, migration, differentiation and apoptosis. Stem cells reside in a defined microenvironment commonly referred to as the stem cell niche to regulate stem cell fate. Given the importance of these environmental cues, there have been numerous tissue engineering attempts to mimic the stem cell niche in vitro to achieve high spatio-temporal control over cell–cell and cell–matrix interactions and reproduce mechano-chemical cues using engineered hydrogels and micro-devices. In 1977, Matrigel, a basement membrane extracellular matrix (ECM) containing a unique mix of ECM components and growth factors, was extracted from mouse sarcoma tumours and used to support in vitro cell culture. Matrigel was later shown to allow breast epithelial cells to grow in three dimensions and form lumens with milk protein secretion6, and adult intestinal stem cells embedded in Matrigel in the presence of a tissue-specific cocktail of growth factors were also shown to self-organize into 3D crypt–villus structures. Organoid research intertwined with 3D cell culture, stem cell and tissue engineering for over a century, with various debates on the definition, standard and scope. An organoid is a self-organized 3D tissue that is typically derived from stem cells (pluripotent, fetal or adult), and which mimics the key functional, structural and biological complexity of an organ. Cells comprising organoids can be derived from induced pluripotent stem cells (iPSCs) or tissue-derived cells (TDCs), including normal stem/ progenitor cells, differentiated cells and cancer cells. Compared with conventional 2D cultures and animal models, organoid cultures enable patient specificity in the model while recapitulating in vivo tissue-like structures and functions in vitro. Organoid cultures are more accessible for manipulation and in-depth biological studies than animal models. As such, organoid cultures have been leveraged for a wide variety of applications including drug discovery, personalized companion diagnostics15 and cell therapy. Moving forward, the trend is to develop more complex models that recapitulate in vivo structure and function as faithfully as possible, in terms of the recapitulating cell types over time, tissue architecture, measurable molecular events and phenotypic functions. Rather than focusing exclusively on the most prominent markers or functional assays, architectural benchmarking of native tissue should also be performed. In the example of hepatocyte organoids, hepatocyte functions are preserved, yet the liver tissue architecture does not match the native tissue where hepatocytes are arranged in cords. Similarly, organoids such as pancreatic or colon cancer organoids grow isotropically, forming a cyst instead of the tubular structure they would form in their native tissue. To derive more complex functions, organoids with multicellular and multi-tissue structures will be important, especially in the context of studying cell–cell interactions. Along this vein, assembloids and organs-on-chips are also becoming increasingly complex and more broadly adopted. On the other hand, the engineer’s approach is to pursue simpler reductionist models defined by the minimal functional modules that drive a complex cellular or tissue function of interest, to study the mechano-biological causation in development or repair, or to develop a robust system for high-throughput screening. The basic premise is that a complex biological function is executed by coordinated operation of a limited number of functional modules, each described by a small set of molecules, and chemical reactions driving physical attribute changes of mesoscale (subcellular or intercellular tissue/multicellular) structures associated with functions of interest in the specific spatio-temporal stage/phase/step. For example, bile canaliculi in the liver exhibit hourly cycles of expansion and contraction. To study the causative contraction events with high resolution, only regions of adjacent hepatocytes forming the bile canaliculi are directly studied in the context of the entire regulatory machinery of adjacent hepatocytes. One can choose to create a much larger structure involving cholangiocytes than the one operated by the minimal functional modules but the model will be noisy and costly. Each functional module is coupled to another and can be modelled together or independently at different length scale. Simple reductionist models have been useful for high-resolution mechanistic understanding of tissue morphogenetic events such as defects. Geometrically constraining the size of the initial 2D seeding pattern and 3D formation by micropatterning and supporting 3D cell growth using Matrigel enabled the induction of tissue-like neural tube morphogenesis and production of highly reproducible neural tubes. This also allowed identification of the mechanisms of neural tube folding and modelled neural tube defects. In another example, symmetry breaking in a uniform sphere of cells and the emergence of a Paneth cell is a critical event in the early stage of intestinal organoid formation. The mechanism was recently elucidated: symmetry breaking is caused by transient activation of the mechanotransducer YAP1, which induces NOTCH-DLL1 lateral inhibition events. YAP1 activation has since been controlled by applying geometrical constraints in hydrogel scaffolds and producing a uniform and reproducible intestinal microtissue3. Organoids can be constrained by reducing the third dimension in a 2.5D culture. The 2.5D culture reduces the depth-driven variabilities of a typical organoid: diffusional constraints in the hypoxic core, limited accessibility for drugs/transfection agents and impeded imaging transparency. Typical examples of restrictions in the third dimension include culturing cells on curved or patterned surfaces, a flattened or constrained cellular construct and overlaying the ECM on a flat cell monolayer at high confluency, which would pull the cells upward, forcing more cell–cell interactions to adopt 3D cell morphology. Hepatocytes in a collagen sandwich have sufficient contact area to attain polarity and form a bile canalicular lumen that contracts in the same periodic cycles as in vivo without the 3D network, wider and cholestatic compared with the native tissue. This cell-based model enables high resolution dissection of the mechanism of bile canaliculi contraction into steps and an understanding of the molecular machinery regulating the phase transitions. We would also envision more engineered organoid models based on CRISPR-edited cells for disease modelling, although these cells and models are synthetic. Besides technological advances in creating more physiologically relevant, robust and easier to use organoid models, we envision seeing greater impact in applications. In the past two decades, although there have been discussions to replace animal testing, these efforts have not yet led to concrete actions. However, this is rapidly changing, with regulatory hard stops now established on multiple fronts. Organoids that can recapitulate the complex physiological functions in vivo have also boosted confidence that the new alternative methods are now viable options. There will be more extrapolation of animal research findings in human organoids to better understand human biology and pathophysiology. We envision widespread adoption of organoids as cell sources for cell therapy, regenerative medicine, in vitro diagnostics and drug discovery. |